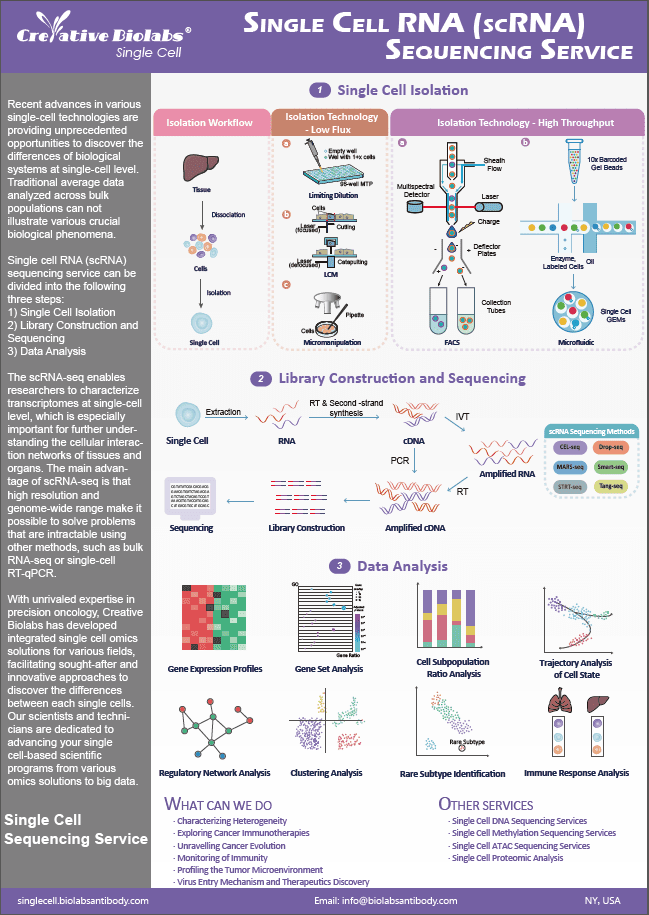
LncRNA Array Service
LncRNA (Long Noncoding RNA) array is a method for profiling lncRNAs in addition to the complete collection of protein-coding mRNA. It is the most effective method for profiling lncRNAs, surpassing many of the limitations of RNA-seq for lncRNAs that are frequently of low abundance.
Overview of LncRNA
Long non-coding RNAs (long ncRNAs, lncRNAs) are a form of RNA that are commonly characterized as transcripts containing more than 200 nucleotides that are not translated into protein. This artificial boundary separates long ncRNAs from tiny non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. LncRNAs consist of intergenic lincRNAs (Long Intervening NonCoding RNAs), intronic ncRNAs, and sense and antisense lncRNAs, with each kind occupying a distinct genomic location in relation to genes and exons.
 Fig. 1 Different types of long non-coding RNAs.1, 3
Fig. 1 Different types of long non-coding RNAs.1, 3
Functions of LncRNA
Despite mounting evidence indicating the vast majority of lncRNAs in mammals are likely to be functional, only a tiny percentage have been proven to be biologically significant. lncRNAs are widely documented to be involved in transcriptional control, epigenetic regulation, DNA replication timing, chromosome stability, and so forth.
Published Data
| Paper Title | Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy |
| Journal | Cell Death & Differentiation |
| Published | 2021 |
| Abstract | Proliferative vitreoretinopathy (PVR) causes severe blindness by forming contractile fibrotic subretinal or epiretinal membranes. PVR is characterized by retinal pigment epithelial (RPE) cell epithelial–mesenchymal transition (EMT). Here, EMT-related lncRNA in RPE (ERLR, LINC01705-201 (ENST00000438158.1)) and its mechanisms are investigated. In RPE cells stimulated with TGF-β1, lncRNA microarray, and RT-PCR showed ERLR is upregulated. Full-length ERLR is mostly expressed in the cytoplasm, according to further studies. In vitro, RPE cells overexpressing ERLR directly induced EMT, while silencing ERLR reduced TGF-β1-induced EMT. Inhibiting ERLR in RPE cells reduced experimental PVR in vivo. Chromatin immunoprecipitation (ChIP) assays showed that TCF4 (Transcription Factor 4) directly binds to ERLR's promoter region and promotes transcription. ERLR directly stabilizes MYH9 protein to induce EMT. TGF-β1 induces EMT in RPE cells via TCF4 and MYH9. RPE cells incubated with vitreous PVR samples also have increased ERLR. ERLR and pan-cytokeratin were colocalized in clinical PVR membrane samples using fluorescent in situ hybridization (FISH) (pan-CK). These findings showed that lncRNA ERLR is involved in PVR and TGF-β1-induced EMT of human RPE cells. This discovery sheds light on PVR's cause and treatment. |
| Result |
Using a lncRNA microarray, differentially expressed lncRNAs in RPE cells stimulated with TGF-β1 were identified. 525 lncRNAs exhibited significant changes in response to TGF-β1 treatment. 133 were upregulated and 392 were downregulated among these lncRNAs. The microarray data were deposited into the GEO database under the accession number GSE105053. Using the microarray results, they chose the top 20 most upregulated lncRNAs and confirmed their expression by RT-PCR in phPRE cells treated with TGF-β1. All 20 lncRNAs were found to be upregulated. lncRNA-LINC01705-201 (ENSG00000232679.2, ENST00000438158.1) was among the top three upregulated lncRNAs with the highest expression abundance and was therefore chosen for further study. TGF-β1 increased the expression of LINC01705-201 significantly in both ARPE-19 and phRPE cells. They hypothesized that this lncRNA may play a crucial role in RPE cell EMT. Consequently, they dubbed it EMT-related lncRNA in RPE cells (ERLR).
|
LncRNA Array Services at Creative Biolabs
In contrast to protein-coding genes, publically accessible lncRNAs are frequently poorly annotated, limited in scope, and dispersed in the collection. Creative Biolabs maintains a high-quality, proprietary lncRNA transcriptome database that collects lncRNAs from all major public databases and repositories, knowledge-based mining of scientific papers, and our lncRNA discovery workflows.
The lncRNA arrays from Creative Biolabs are designed to profile lncRNAs in addition to all protein-coding mRNAs. They are the most effective method for profiling lncRNAs, addressing many of the limitations of RNA-seq for lncRNAs that are frequently rare. For more information, please contact us.
Features & Benefits
-
High Sensitivity and Specificity
The microarray technology we use is highly sensitive and specific, enabling the detection of low-abundance lncRNAs with great accuracy. This is particularly beneficial for studying lncRNAs that are expressed at low levels but play crucial regulatory roles.
-
Comprehensive Coverage
Our arrays cover thousands of lncRNAs and mRNAs, providing a broad view of gene expression profiles. This comprehensive coverage allows for the identification and study of a wide range of lncRNAs, including those with low expression levels, which are often missed by RNA-seq.
-
Quality Control and Data Analysis
Our service includes rigorous quality control steps and in-depth data analysis, ensuring that you receive reliable and high-quality results. This includes RNA isolation, cDNA synthesis, target preparation, array hybridization, and data extraction and summarization.
-
Robust and Mature Technology
Microarray technology is well-established and mature, providing consistent and reproducible results. This reliability makes it a preferred choice for many researchers in the field.
-
Cost-Effective
Compared to RNA-seq, our LncRNA Array Service is more cost-effective, especially when it comes to profiling low-abundance lncRNAs. This makes it an economical choice for large-scale studies.
Q&As
Q: What are the benefits of using LncRNA arrays?
A: Benefits include comprehensive coverage of full-length lncRNAs, systematic annotation, high sensitivity, and a robust collection of lncRNAs from major databases. These arrays are particularly useful for identifying and quantifying low-abundance lncRNAs.
Q: What types of lncRNAs can be profiled using this service?
A: The service profiles various types of lncRNAs, including intergenic lincRNAs, intronic ncRNAs, and sense and antisense lncRNAs. Each type occupies distinct genomic locations relative to genes and exons.
Q: How does the LncRNA array handle low-abundance lncRNAs?
A: LncRNA arrays use sequence-specific probes that enable accurate detection and quantification of low-abundance lncRNAs, which are often missed by RNA-seq due to competition with high-abundance transcripts.
Q: What is the sample requirement for LncRNA array analysis?
A: Typically, total RNA extracted from the sample of interest is required. The quality and quantity of RNA are critical for the successful performance of the array.
Q: How does the LncRNA array ensure data accuracy and reliability?
A: The LncRNA array uses multiple probes per transcript, rigorous quality control steps, and sophisticated data analysis methods to ensure high accuracy and reliability. These arrays are designed to minimize cross-hybridization and maximize specificity.
Resources
References
- Fernandes, Juliane C R et al. "Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease." Non-coding RNA vol. 5,1 17. 17 Feb. 2019, doi:10.3390/ncrna5010017.
- Yang, Shuai et al. "Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy." Cell death and differentiation vol. 28,8 (2021): 2351-2366. doi:10.1038/s41418-021-00756-5
- Distributed under Open Access license CC BY 4.0, without modification.
Search...


 Fig. 2 LncRNA expression profiles are altered in RPE cells treated with TGF-β1.2, 3
Fig. 2 LncRNA expression profiles are altered in RPE cells treated with TGF-β1.2, 3