- Home
- Single Cell Research Area
- Single Cell Omics Solutions for Oncology
- Single Cell Omics Solutions for Cancer Stem Cells
Single Cell Omics Solutions for Cancer Stem Cells
Single-cell sequencing refers to the technique of obtaining corresponding data and performing information analysis by sequencing a single cell at the genome, transcriptome, and epigenome levels. Single-cell sequencing technology is a powerful tool that has attracted much attention in recent years. It can comprehensively analyze cell heterogeneity in the same kind of stem cells and identify cells with different phenotypes. Creative Biolabs has extensive research experiences in single-cell sequencing, which covers single-cell genome sequencing, single-cell exome sequencing, single-cell targeted sequencing, single-cell RNA sequencing, single-cell methyl sequencing, and single-cell proteomic analysis, etc. With our advanced platforms, we offer comprehensive single cell omics solutions for cancer stem cells.
scWGS for Cancer Stem Cells
Single-cell whole-genome sequencing (scWGS) is a novel technology for amplification and sequencing of whole genomes at the single-cell level. Single Cell offers advanced scWGS services for our clients to facilitate your cancer stem cell research at the single cell level. Our technology can efficiently amplify trace whole genome DNA, identify high coverage complete genome, perform high-throughput sequencing in cancer stem cells, which play an important role in obtaining genetic variation information and revealing stem cell differences for cancer therapy.
scWES for Cancer Stem Cells
Single-cell exome sequencing technology (scWES) is a genomic analysis method that amplifies genomic DNA at single cell level, captures and enriches the DNA of the whole genome exome region. Single Cell has delivered numerous scWES services to our worldwide customers. Through our platform, we will help our client with the perfect solutions for detecting the exome sequence, the expression level of specific cancer stem cells. We will be always dedicated to accelerating your project and getting meaningful data in a cost-effective manner.
scTargeted-seq for Cancer Stem Cells
Single cell targeted sequencing (scTargeted-seq) is a high-throughput sequencing technology that is mainly used for mutational analysis of tumor stem cells. Currently, Single Cell has established a unique scTargeted-seq platform to help determine the disease-causing genes and susceptibility genes without causing many mutated genes in different types of cancer stem cells. The results can be highly accurate and reproducible, as well as can be accepted by authorities.

 Fig.2 Schematic overview of scRNA-seq. 2
Fig.2 Schematic overview of scRNA-seq. 2
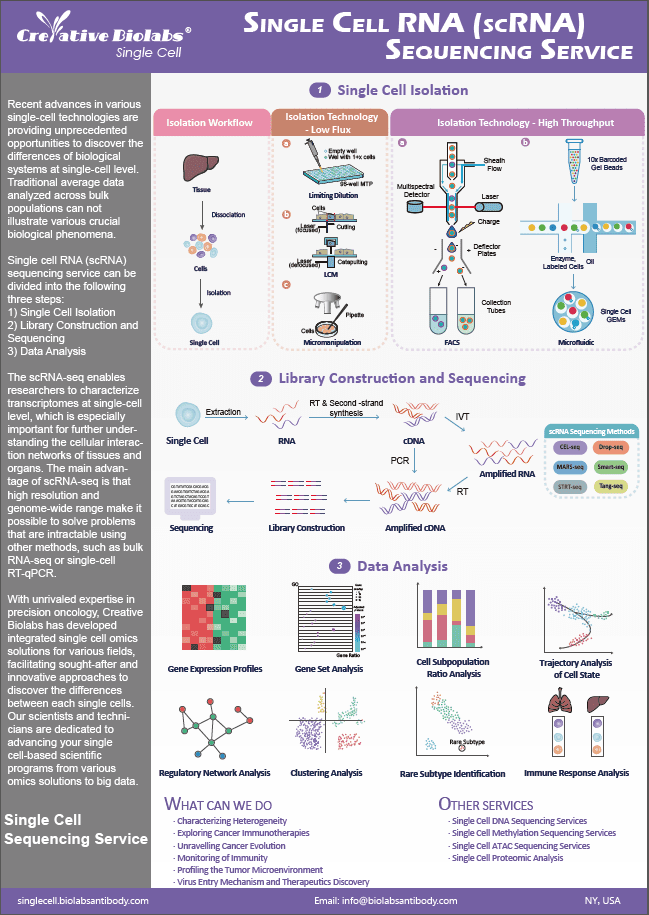
scRNA-seq for Cancer Stem Cells
Single cell RNA sequencing (scRNA-seq) has been considered as an attractive tool for revealing cancer stem cells at a single-cell level. In Single Cell, we offer a number of scRNA-seq services to study intra-tumor heterogeneity of cancer stem cells and their functions in cancer occurrence, development and treatment. Our scRNA-seq assays can be widely used for identifying in vivo genomic alterations of cancer stem cells. Recently, we have successfully generated a combination assay based on scRNA-seq and DNA sequencing to assess the gene expression in various cancer stem cells.
scMethyl-Seq for Cancer Stem Cells
Single cell methylation sequencing (scMethyl-Seq) is a stable system for identifying cell types and factors that affect cell functions. It plays a vital role in maintaining normal cell function, transmitting genomic imprinting, embryonic development, and tumorigenesis. Equipped with advanced technologies, we are committed to providing high-quality scMethyl-Seq services to map single-base resolution DNA methylation profiles. In general, our quantitative scMethyl-Seq assays can be broadly used for revealing methylation patterns in cancers, testing unique expression level of individual cells, and assessing tumor burden in different cell types, such as stem cells.
 Fig.3 Schematic overview of scProteomic Analysis.3, 4
Fig.3 Schematic overview of scProteomic Analysis.3, 4
scProteomic Analysis for Cancer Stem Cells
Single-cell proteomic technologies can provide a comprehensive profiling of protein levels in a variety of single cells. Our expert team has developed a number of scProteomic analysis assays to assess the interaction between cancer stem cells, the functional of the immune cells, as well as the drug efficacy for cancer therapy. These assays are mainly based on flow cytometry (FCM), enzyme-linked immunospot, microchips, as well as DNA barcoding technology.
Our Highlights
- Rich project experience
- Leading database technology
- Customized service
- Short project cycle
Creative Biolabs has the most authoritative testing strategies and tools to provide you with single-cell omics services. For more detailed information, please feel free to contact us or directly sent us an inquiry.
Q&As
Q: How can single-cell omics solutions aid in the development of cancer therapies?
A: Single-cell omics solutions enable the identification of unique molecular characteristics and therapeutic targets within cancer stem cells. This knowledge helps in designing targeted therapies that can effectively eliminate cancer stem cells, potentially preventing tumor recurrence and improving patient outcomes.
Q: What is the significance of studying cellular heterogeneity in cancer stem cells?
A: Studying cellular heterogeneity in cancer stem cells is crucial for understanding tumor diversity and the mechanisms driving cancer progression and resistance to treatments. Single-cell omics technologies provide detailed insights into this heterogeneity, facilitating the development of more effective and personalized therapeutic strategies.
Q: How do single-cell multi-omics approaches enhance cancer stem cell research?
A: Single-cell multi-omics approaches integrate data from various omics platforms, such as genomics, transcriptomics, proteomics, and metabolomics. This comprehensive analysis provides a holistic view of cancer stem cell biology, enabling the identification of complex regulatory networks and interactions between different molecular layers.
Q: What are the challenges of single-cell omics in cancer stem cell research?
A: Challenges include technical complexities in isolating and analyzing single cells, data integration from multiple omics platforms, and the high cost of these advanced technologies. Overcoming these challenges requires continuous technological advancements and interdisciplinary collaboration.
Q: What future advancements are expected in single-cell omics for cancer stem cells?
A: Future advancements may include improved techniques for single-cell isolation and sequencing, enhanced data integration methods, and the development of more cost-effective solutions. These advancements will likely lead to deeper insights into cancer stem cell biology and the discovery of novel therapeutic targets.
Resources
References
- Rodriguez-Meira, Alba et al. "Unravelling Intratumoral Heterogeneity through High-Sensitivity Single-Cell Mutational Analysis and Parallel RNA Sequencing." Molecular cell vol. 73,6 (2019): 1292-1305.e8. doi: 10.1016/j.molcel.2019.01.009.
- Hwang, Byungjin et al. "Single-cell RNA sequencing technologies and bioinformatics pipelines." Experimental & molecular medicine vol. 50,8 1-14. 7 Aug. 2018, doi:10.1038/s12276-018-0071-8. Distributed under Open Access license CC BY 4.0. The image was modified by extracting and using g part of the original image.
- El Kennani, Sara et al. "Proteomic Analysis of Histone Variants and Their PTMs: Strategies and Pitfalls." Proteomes vol. 6,3 29. 21 Jun. 2018, doi:10.3390/proteomes6030029.
- Distributed under Open Access license CC BY 4.0, without modification.
Search...