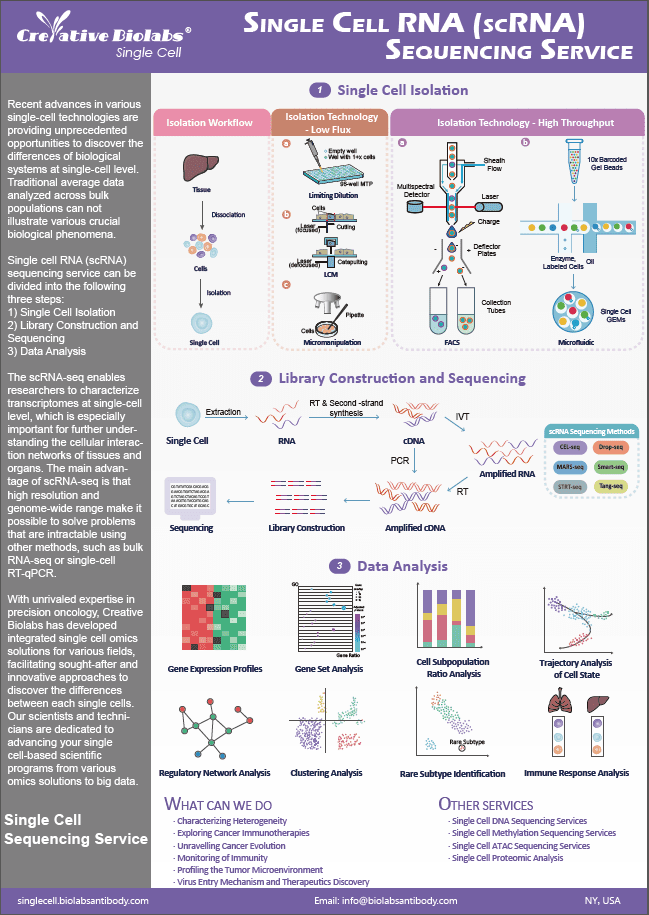
Single Cell Spatial Gene Expression Services
Creative Biolabs provides a comprehensive range of customized, high-quality services in single cell spatial gene expression service to help researchers map the whole transcriptome with morphological context in FFPE or fresh-frozen tissues to discover novel insights into normal development, disease pathology, and clinical translational research.
Single Cell Spatial Gene Expression Technology Principle
Spatial transcriptome technology utilizes the 10x Genomics Visium platform, which uses two kinds of slides, tissue permeabilization slide and gene expression slide. First, the tissue permeabilization slide is used to explore the permeabilization conditions of the target area of the tissue section. After determining the optimal conditions for permeabilization, gene expression slide is utilized to capture RNA and library construction of tissue sections. The visium spatial gene expression slide has 4 capture areas (6.5 x 6.5 mm), each defined by a fiducial frame (fiducial frame + Capture Area is 8 x 8 mm). The capture area has ~5,000 gene expression spots, each spot has a unique barcode sequence for position mapping.
Tissue sections placed on these capture areas are fixed and stained, permeabilized, and cellular mRNA is captured by the primers on the gene expression spots. All the cDNA generated from mRNA captured by primers on a specific spot share a common Spatial Barcode. Libraries are generated from the cDNA and sequenced and the Spatial Barcodes are used to associate the reads back to the tissue section images for spatial gene expression mapping.
Our Single Cell Spatial Gene Expression Services
Multi-group analysis can quickly collect a significant amount of data. By merging the research findings with the key analysis, the results can be generated rapidly. The data from omics at various levels may be cross-verified, making the experiment's conclusion gained through extensive analysis more credible. Multi-layer omics integration can compensate for the shortcomings of single-layer omics in detecting preference differences. The fault induced by the difference in preference in single group detection can be compensated for by multilayer omics integration. It is simpler to discover the difference by evaluating downstream omics data, as there is a propensity to intensify the influence from gene to protein to metabolism.
We offer scientific and meticulous design for tissue section preparation firstly. Then we use standard fixation and staining techniques to visualize tissue sections on slides. For fresh frozen tissue, the tissue is permeabilized to release mRNA from the cells, for FFPE tissues, tissue is permeabilized to release ligated probe pairs from the cells, which binds to the spatially barcoded oligonucleotides present on the spots. The barcoded cDNA is compatible with standard NGS short-read sequencing on Illumina sequencers for massive transcriptional profiling of entire tissue sections. Finally, we use Space Ranger analysis software to process spatial gene expression data.
 Fig.1 Our Single Cell Spatial Gene Expression Service. (Creative Biolabs)
Fig.1 Our Single Cell Spatial Gene Expression Service. (Creative Biolabs)
Published Data
- Schematic stained cross-section of sample (top). Visualization of colocalized spatial mRNA expression (bottom left). Confocal immunofluorescence image (bottom right).
- Cell cluster.
- Mapping of data points to spatial positions show that clusters are localized to distinct histological regions.
 Fig.2 Heatmap of the 20 most variable genes in different regions.1
Fig.2 Heatmap of the 20 most variable genes in different regions.1
 Fig.3 Enriched pathways analysis.1
Fig.3 Enriched pathways analysis.1
Applications
Single cell spatial gene expression service can help researchers obtain transcriptome data in cells located at different positions of the section from tissue, visual analysis of gene expression and morphological data by dedicated software, and have a wide range of applications in the fields of pathology, neuroscience or developmental biology.
 Fig.7 Applications of single cell spatial gene expression. (Creative Biolabs Original)
Fig.7 Applications of single cell spatial gene expression. (Creative Biolabs Original)
Please contact us for your tailored solution about single cell spatial transcriptome.
Features & Benefits
- Spatial Resolution: Our service offers high spatial resolution, allowing researchers to visualize gene expression patterns within intact tissue sections at single-cell resolution.
- Multiplexed Analysis: With Single Cell Spatial Gene Expression Services, multiple genes can be simultaneously analyzed within the same tissue section, providing a comprehensive view of spatially resolved gene expression profiles.
- Tissue Architecture Preservation: We preserve tissue architecture during the analysis, ensuring that spatial relationships between cells and tissue structures are maintained, facilitating accurate interpretation of gene expression patterns.
- Cell Type Identification: Researchers can identify different cell types within tissue sections based on their spatial gene expression profiles, enabling the study of cell heterogeneity and interactions within the tissue microenvironment.
- Insights into Disease Mechanisms: By elucidating spatial gene expression patterns, researchers can gain insights into disease mechanisms, tissue organization, and cellular responses to stimuli, advancing our understanding of complex biological processes and disease pathogenesis.
FAQs
Q: What is Single Cell Spatial Gene Expression analysis, and how does it differ from traditional single-cell RNA sequencing (scRNA-seq)?
A: Single Cell Spatial Gene Expression analysis involves profiling gene expression within intact tissue sections at single-cell resolution, preserving spatial information. Unlike traditional scRNA-seq, which dissociates cells from tissues, spatial analysis maintains tissue architecture, offering insights into cellular interactions within the tissue microenvironment.
Q: What types of samples are compatible with Single Cell Spatial Gene Expression Services?
A: Our service is compatible with various tissue types, including fresh-frozen or FFPE tissues. Whether it's tumor samples, organ sections, or tissue microarrays, we can analyze spatial gene expression patterns to elucidate biological insights relevant to your research.
Q: How is spatial gene expression data analyzed, and what insights can be gained from the results?
A: Spatial gene expression data are analyzed using computational methods to identify gene expression patterns within tissue sections. Insights gained include the spatial distribution of different cell types, localization of specific gene markers, and understanding tissue organization and cellular interactions in the context of disease pathology or developmental processes.
Q: Can Single Cell Spatial Gene Expression Services identify rare cell populations within tissues?
A: Yes, our service can identify rare cell populations within tissues based on their spatial gene expression profiles. By analyzing intact tissue sections, we can detect and characterize both abundant and rare cell types, providing valuable insights into cellular heterogeneity and rare cell interactions within the tissue microenvironment.
Resources
Reference
- Berglund, Emelie et al. "Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity." Nature communications vol. 9,1 2419. 20 Jun. 2018, doi:10.1038/s41467-018-04724-5. Distributed under Open Access license CC BY 4.0, without modification.
Search...