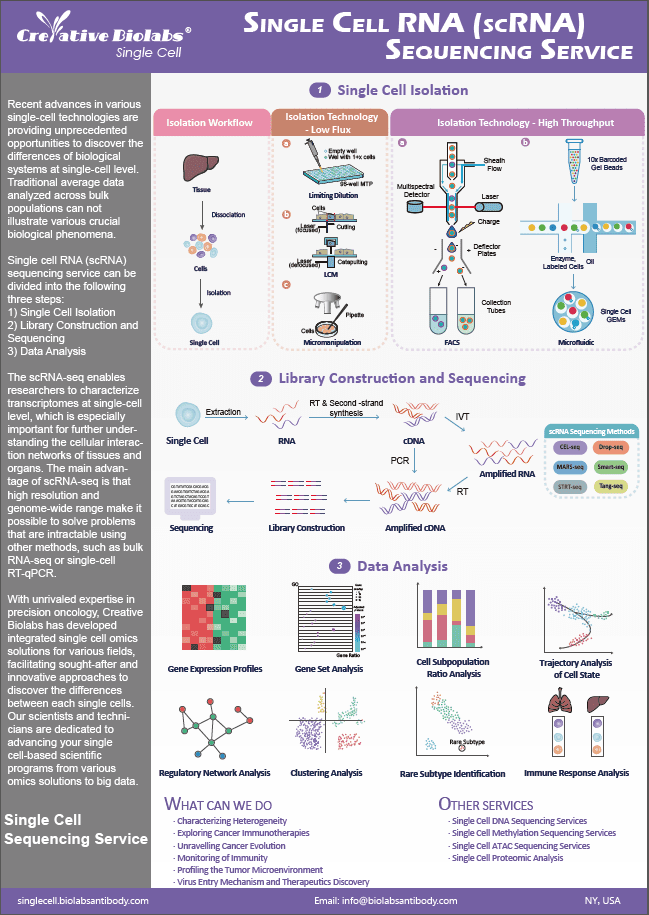
Single Cell CITE-Seq Service
Single cell CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) is a droplet microfluidic method for simultaneously analyzing the transcriptome and proteome of a single cell.
Introduction to Single Cell CITE-Seq
Single cell CITE-seq measures the abundance of their targets using DNA-conjugated antibodies and sequencing. The DNA oligos were also designed to be compatible with existing single cell DNA sequencing. With over 1000 genes and 80 proteins measured from thousands of individual cells; this technique has produced some of the most comprehensive measurements of single cells to data.
Published Data
| Paper Title | Comprehensive integration of single-cell data |
| Journal | Cell |
| Published | 2019 |
| Abstract | Single-cell transcriptomics has revolutionized our capacity to characterize cell states, but profound biological comprehension requires more than a taxonomic listing of clusters. As new methods emerge to measure distinct cellular modalities, integrating these datasets to better comprehend cellular identity and function represents a significant analytical challenge. Here, the authors develop a strategy to "anchor" disparate datasets together, allowing us to integrate single cell measurements not only through scRNA-seq technologies but also across modalities. They anchor scRNA-seq experiments with scATAC-seq to investigate chromatin differences in closely related interneuron subsets and map protein expression measurements onto a bone marrow atlas to characterize lymphocyte populations. Finally, they harmonize in-situ gene expression and scRNA-seq datasets to enable transcriptome-wide imputation of spatial gene expression patterns. Their work presents a strategy for the assembly of standardized references and the transfer of data between datasets. |
| Result | They performed CITE-seq on 33,454 human bone marrow cells, measuring transcriptomes and 25 cell-surface proteins (median 4,575 RNA unique molecular identifiers [UMIs] and 2,312 antibody-derived tags [ADT] UMIs per cell). First, they cross-validated CITE-seq data by randomly assigning cells to a reference or query dataset and identifying anchors. They predicted protein levels in the query dataset using a weighted average of CITE-seq counts from reference anchor cells. For most proteins (23/25), they observed a strong correlation between measured and imputed expression levels (median R=0.826). The remaining residual included background CITE-seq binding (perhaps driven by cell size), stochastic variation in protein expression, or technical noise. Poor antibody specificity or a lack of transcriptomic markers that correlate with immunophenotype could explain poor correlations in two cases. Indeed, CD25 and CD197-CCR7 expression patterns show sporadic ADT binding across all cells, indicating non-specific antibody binding confounding the biological signal. By downsampling RNA features used to identify anchors and repeating cross-validations, they found prediction accuracy saturating at 250–500 features, suggesting only a subset of shared genes need to be measured across experiments to transfer additional modalities across datasets. |
Single Cell CITE-Seq Service
Creative Biolabs offers single cell CITE-seq services, including cell isolation using a droplet system, data generation, and bioinformatics analysis, giving you a comprehensive understanding of the single cell transcriptome and proteome at the same time. For more information, please contact us.
Features & Benefits
-
Simultaneous RNA and Protein Profiling:
Single Cell CITE-Seq enables simultaneous analysis of both RNA and protein expression within individual cells, providing a comprehensive view of cellular phenotypes and functions.
-
Multi-Omics Integration:
By combining RNA and protein analysis, Single Cell CITE-Seq facilitates multi-omics integration, allowing for the correlation of gene expression with protein abundance, leading to deeper insights into cellular biology.
-
High Throughput and Scalability:
Our service offers high throughput and scalability, enabling the analysis of thousands to millions of single cells in a single experiment, facilitating large-scale studies and in-depth exploration of cellular diversity.
-
Fine-Resolution Cellular Characterization:
Single Cell CITE-Seq provides fine-resolution characterization of individual cells, allowing for the identification of rare cell populations, cell states, and transitional cell types within heterogeneous tissues.
FAQs
Q: What types of samples are compatible with Single Cell CITE-Seq Service?
A: Single Cell CITE-Seq Service can be applied to various sample types, including cell suspensions from tissues, biofluids, or cell cultures. Samples should contain a sufficient number of viable cells for analysis.
Q: How does Single Cell CITE-Seq Service handle cell multiplets or doublets?
A: Single Cell CITE-Seq Service employs bioinformatics methods to identify and remove cell multiplets or doublets during data analysis. This ensures that only single-cell data are used for downstream analysis, minimizing potential confounding effects.
Q: What insights can be gained from Single Cell CITE-Seq data?
A: Single Cell CITE-Seq data can provide insights into cell heterogeneity, cellular interactions, signaling pathways, and biomarker discovery. Researchers can use this information to study disease mechanisms, identify therapeutic targets, and evaluate treatment responses.
Q: How can researchers analyze and interpret Single Cell CITE-Seq data?
A: Analysis of Single Cell CITE-Seq data involves bioinformatics processing to quantify gene expression and protein levels in individual cells. Researchers can perform clustering analysis, differential expression analysis, and pathway enrichment analysis to interpret the data and extract meaningful biological insights.
Resources
Search...