Single Cell ATAC Service
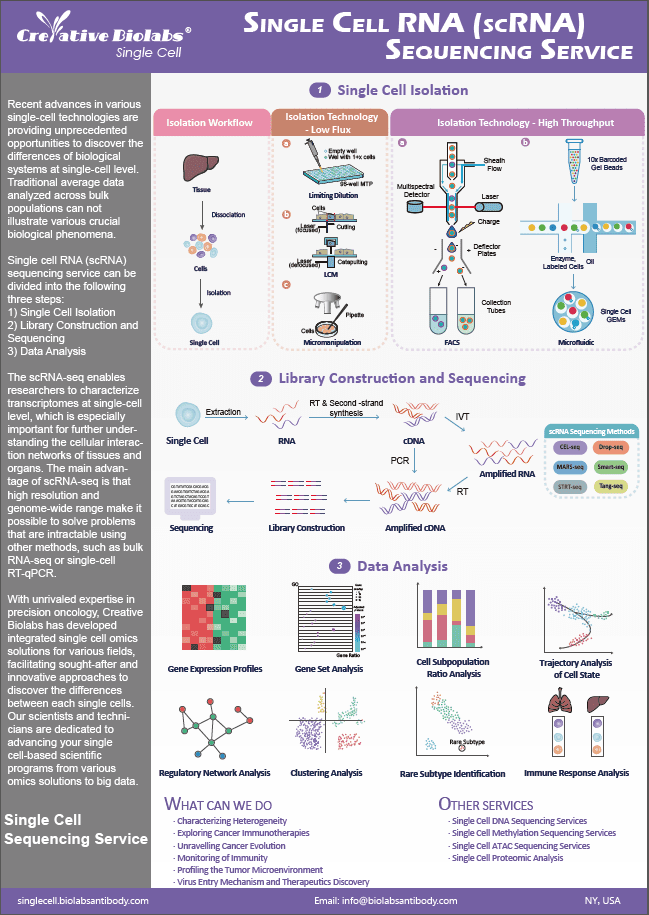
Creative Biolabs provides a comprehensive range of customized, high-quality services in single cell ATAC profiling to support various scientific researches worldwide. Analyzing chromatin accessibility at the single cell level can be used to provide insights into gene regulatory mechanisms.
ATAC
Eukaryotic chromosomes are formed by a series of folding events in nucleosome structures generated by DNA molecules wrapping histone proteins. There are some regions of chromatin that are loosely reformed, called open chromatin or open chromatin regions, where both DNA replication and gene transcription occur. Therefore, obtaining the information will help us understand the regulation of gene expression by open regions, as well as better understanding of biological regulation.
ATAC-seq (Assay for transposase-accessible chromatin with high-throughput sequencing) is a rapid and sensitive epigenetic research technique that uses the Tn5 transposase to cleave open chromatin regions, coupled with sequencing primers (adapter) for high-throughput sequencing (Buenrostro; et al., 2013).
Single Cell ATAC Technology
The 10x Genomics single cell ATAC technology enables the detection of open regions of cellular chromatin at the single cell level. First, the transposase enters the nucleus and preferentially cleaves DNA in the open chromatin regions, while adding sequencing primers to the ends of the DNA fragments; second, in the oil droplets the gel beads with barcodes carry out barcode labeling on the DNA fragments in each cell, and the DNA with barcode labels is used for further library construction; finally, sequencing the library on the Illumina platform, a large number of single cell DNA chromatin open region data can be obtained in one experiment.
Our Single Cell ATAC Service
We offer scientific and meticulous design for cell suspension, library construction, sequencing and data analysis to ensure high-quality research results. If you want to simultaneously analyze epigenomic landscape and gene expression in the same single cell, please visit Single Cell Gene Expression and ATAC Service for further information.
 Fig.1 Our Single Cell ATAC Service. (Creative Biolabs)
Fig.1 Our Single Cell ATAC Service. (Creative Biolabs)
Features & Benefits
1. High throughput
About 500-10,000 nuclei can be obtained per channel efficiently; for high throughput platform, we can run 8 samples (80,000 nuclei per run) in one experiment.
2. High resolution
The chromatin accessibility information can be obtained at a single-cell nucleus level.
3. High capture efficiency
The capture efficiency of nuclei can reach up to 65% per sample.
4. Ultra-low contamination
The number of mitochondrial DNA reads is less than 2%.
5. More comprehensive information
We can get information on all open chromatin regions, not limited to a single transcription factor.
6. One-stop service
We can provide one-stop service from sample processing to data visualization; or provide customized analysis procedures with the most up-to-date technical solutions.
Results Display
Here are some results displayed that refers to the drawings of single cell ATAC profiling articles.
1. Rare cell detection through scATAC-seq.
2. Analyzed TF activity and found distinct patterns of activity in different cell types.
3. Dynamic chromatin changes visualization through motif locus.
 Fig.2 Motif locus analysis map. 1
Fig.2 Motif locus analysis map. 1
Application Prospect
Single cell gene ATAC service can help researchers get insight into stem cell development, mechanisms of gene regulation, and have a wide range of applications in the fields of neuroscience or immune research.
 Fig.3 Applications of single cell ATAC. (Creative Biolabs)
Fig.3 Applications of single cell ATAC. (Creative Biolabs)
If you are interested in the single cell ATAC service, please contact us to learn more. Our scientists will reply within 24 hours.
Q&As
Q: How does Single Cell ATAC-Seq differ from bulk ATAC-Seq?
A: Unlike bulk ATAC-Seq, which provides an average signal from a mixture of cells, Single Cell ATAC-Seq offers insights at the resolution of individual cells. This allows for the detection of cell-specific regulatory elements and the study of cellular heterogeneity within a population.
Q: What are the key steps in the Single Cell ATAC-Seq workflow?
A: The workflow involves nuclei isolation, transposition of DNA using Tn5 transposase, partitioning of nuclei into nanoliter-scale Gel Beads-in-emulsion (GEMs), PCR amplification, and sequencing. This is followed by bioinformatic analysis to map and quantify open chromatin regions.
Q: What bioinformatic tools are used for Single Cell ATAC-Seq data analysis?
A: Data analysis typically involves using tools such as Cell Ranger ATAC for primary and secondary analysis, and Loupe Browser for interactive data visualization and exploration. These tools help in identifying open chromatin regions, performing differential accessibility analysis, and annotating motifs.
Q: What sample types can be used for Single Cell ATAC-Seq?
A: Single Cell ATAC-Seq can be performed on a variety of sample types, including primary cells, cultured cell lines, and tissue samples. Proper nuclei isolation and preparation are crucial for obtaining high-quality data from different sample sources.
Q: What support is available for implementing Single Cell ATAC-Seq in research?
A: Comprehensive support includes protocols for sample preparation, guidelines for data analysis, and access to bioinformatic tools. Many service providers offer consultation and technical assistance to ensure successful implementation and accurate data interpretation.
Resources
Reference
- Bravo González-Blas, Carmen et al. "Identification of genomic enhancers through spatial integration of single-cell transcriptomics and epigenomics." Molecular systems biology vol. 16,5 (2020): e9438. doi:10.15252/msb.20209438. Distributed under Open Access license CC BY 4.0, without modification.
Search...