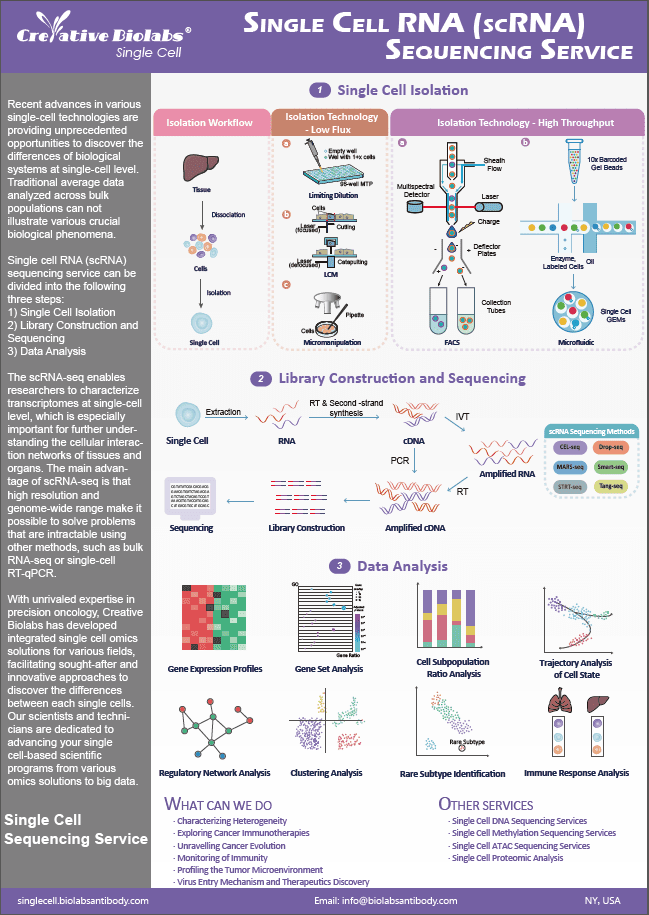
Single Cell ChIP-Seq Service
Single cell ChIP-Seq (chromatin immunoprecipitation followed by sequencing) is a method for mapping histone modifications, transcription factors, and other protein-DNA interactions in a single cell genome-wide.
A limitation of chromatin mapping technologies is that they require large amounts of input material and yield 'average' profiles insensitive to cellular heterogeneity. This is a significant shortcoming, given that cell-to-cell variability is inherent to most tissues and cell populations.
Single cell ChIP-Seq (scChIP-seq)
To study the intra-tumor heterogeneity of chromatin states, scientists developed a high-throughput scChIP-seq approach that combines droplet microfluidics with single cell DNA barcoding technologies. This scChIP-seq approach enables the segmentation of cell populations solely based on their chromatin landscape and the identification of key chromatin features of each subpopulation.
scChIP-seq Workflow
Published Data
| Paper Title | Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state |
| Journal | Nature Biotechnology |
| IF | 54.908 |
| Published | 2019 |
| Abstract | A flexible way to research functional genomic components and their control is through chromatin profiling. However, the ensemble profiles produced by the present techniques are not sensitive to cell-to-cell variance. Here, authors integrate sequencing, DNA barcoding, and microfluidics to gather chromatin data at the single-cell level. By testing tens of thousands of individual cells, they show how useful the technology is by using the information to create high-quality chromatin state maps for each cell type from a mixture of fibroblasts, hematopoietic progenitors, and embryonic stem cells (ES cells). Each cell only contains a small amount of data, roughly 1,000 distinct reads. Nevertheless, by analyzing a large number of ES cells, they distinguish a variety of subpopulations based on variations in the chromatin signatures of pluripotency and differentiation priming. By comparing these results to orthogonal single-cell gene expression data, they confirm these findings. Single-cell analysis using new techniques shows the epigenetic heterogeneity of transcriptional analysis. |
| Results | They determined the number of reads that overlapped each signature for each individual ES cell (or mouse embryonic fibroblasts, MEFs), resulting in a matrix of 5,405 single cells with 91 signatures. The signature matrix's aggregative hierarchical clustering identified several notable cell populations with correlated chromatin landscapes. All MEFs were separated from ES cells, which were dispersed throughout various clusters, by the major division. They created multi-dimensional scaling (MDS) plots from the signature matrix to show the relationship between cells. The MEFs' tighter distribution is indicative of H3K4me2 landscapes that are more consistent. In contrast, the MDS plot shows that the individual ES cells cover a much larger area, dividing into three loose groups. The observation that such lineage-committed cells adopt a relatively constrained chromatin state may explain the tighter distribution among specific MEFs. In contrast, the accessible and malleable state of the chromatin. |
At Creative Biolabs, we offer scChIP-seq for a better interpretation of the intra-tumor heterogeneity of chromatin states. If you have any requirements, please feel free to contact us for further communication about your project.
Features & Benefits
-
High-Resolution Profiling
Single Cell ChIP-Seq enables high-resolution mapping of protein-DNA interactions down to the level of individual cells, providing insights into cellular heterogeneity that is often missed in bulk assays. -
Enhanced Sensitivity
The service uses advanced technologies that require less input material than traditional ChIP-Seq, making it possible to study samples where cell numbers are limited. -
Comprehensive Chromatin Analysis
This service allows for the detailed examination of histone modifications and transcription factor bindings across the genome, helping to elucidate complex regulatory networks. -
Droplet Microfluidics Integration
Incorporating droplet microfluidics with single-cell DNA barcoding significantly improves the throughput and precision of chromatin state analyses at the single-cell level. -
Tailored for Heterogeneity
Single Cell ChIP-Seq is particularly suited for studying intra-tumor heterogeneity and other complex tissue environments, providing a deeper understanding of disease mechanisms.
Q&As
Q: What sample types are required for Single Cell ChIP-Seq?
A: Single Cell ChIP-Seq can be performed on various sample types, including freshly isolated cells, cryopreserved cells, and even challenging samples like FFPE tissues, with appropriate pre-treatment and quality control steps.
Q: How is the data from Single Cell ChIP-Seq analyzed?
A: Data analysis in Single Cell ChIP-Seq involves aligning sequencing reads to a reference genome, identifying peaks corresponding to protein-DNA interactions, and clustering cells based on their chromatin landscapes. Advanced bioinformatics tools are used to interpret the data and provide insights into cell-specific regulatory mechanisms.
Q: How scalable is Single Cell ChIP-Seq for large-scale studies?
A: Single Cell ChIP-Seq is becoming increasingly scalable with advancements in automation and high-throughput sequencing technologies. While it traditionally required extensive manual handling, new methods and platforms are streamlining the process, enabling large-scale studies involving thousands of individual cells.
Q: What support services are available for Single Cell ChIP-Seq?
A: Many companies and academic centers offer support services for Single Cell ChIP-Seq, including experimental design, sample preparation, sequencing, and data analysis. These services are invaluable for researchers new to the field or those without access to the necessary equipment.
Q: What are the quality control measures in Single Cell ChIP-Seq experiments?
A: Quality control measures in Single Cell ChIP-Seq experiments include assessing the efficiency of chromatin immunoprecipitation, evaluating the purity and integrity of DNA, and verifying the sequencing quality. Proper controls, such as input DNA and negative immunoprecipitation controls, are also critical to ensure the reliability of the data.
Resources
Search...