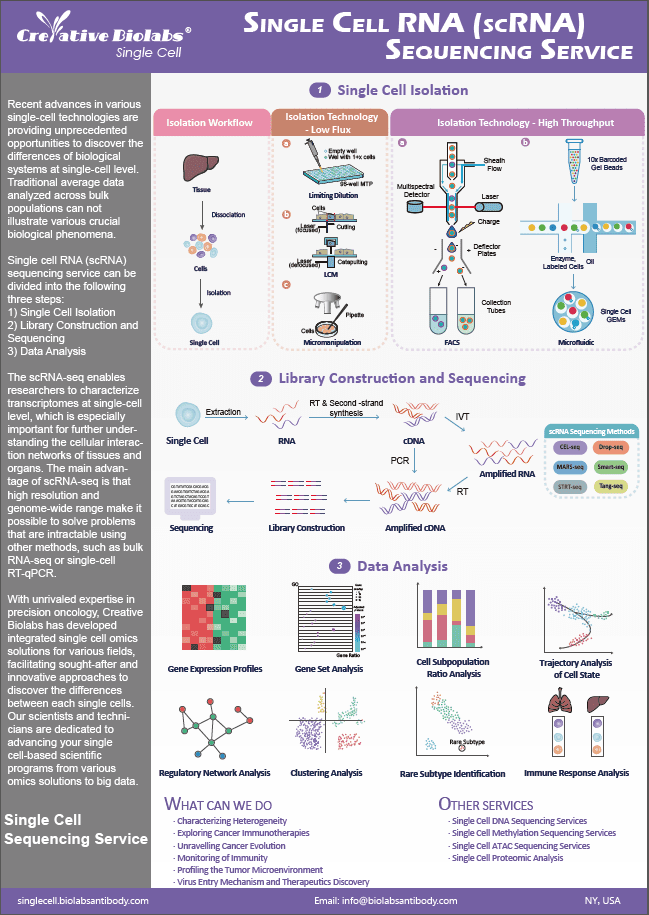
Spatial Metabolomics Service
By using the Mass Spectrometry Imaging (MSI) instrument, metabolites on tissue sections are collected and detected point by point. Through data analysis, the metabolites detected at each point on the tissue section are restored to a two-dimensional plane, thereby obtaining qualitative, quantitative, and localization information of small molecule metabolites.
Introduction of AFADESI-MSI
Creative Biolabs has made significant advancements in their spatial metabolomics platform through the integration of desorption electrospray ionization mass spectrometry imaging (DESI-MSI) technology, resulting in the pioneering development of air flow-assisted ionization desorption electrospray ionization mass spectrometry imaging (AFAI-MSI) technology. By employing airflow and transmission tubes for the extended conveyance of charged droplets, these droplets undergo further desolvation, enrichment, and ionization through the influence of high-speed airflow and voltage. This remarkable enhancement elevates the sensitivity of detection to unprecedented levels. Furthermore, it expands the spatial and operational versatility of the samples under examination, enabling the collection and imaging analysis of voluminous samples over long distances, transcending the limitations of solely single tissue samples.
Workflow of Spatial Metabolomics (AFADESI-MSI)
1. A certain high voltage is applied to the electrospray capillary nozzle. The spray solvent flows out from the inner tube of the nebulizer and is quickly atomized into charged spray droplets by the high-pressure nitrogen ejected from the outer tube.
2. The high-speed charged droplets that are sprayed out bombard the surface of the sample to be tested, and the sample is simultaneously desorbed and ionized under the effect of solvent extraction.
3. The charged droplets containing the sample to be tested rapidly undergo desolvation. They enter the analyzer through the collection cone hole of the mass spectrometer and are detected.
4. The sample is fixed on the carrying platform, and the carrying platform is moved continuously or in pulses to perform a two-dimensional scan of the sample.
5. Meanwhile, the mass spectrometer records the intensity of the mass spectral signal to obtain the molecules on the sample surface and their content.
6. After this information is converted through mass spectrometric imaging analysis software, a two-dimensional spatial intensity distribution map of the selected ions or ion groups can be obtained.
Single Cell Spatial Metabolomics Service
At Creative Biolabs, we can provide highly customized spatial metabolomics service. We are aiming to help you elucidating the cellular heterogeneity of metabolite generation under particular conditions at particular time intervals.
For more information, please contact us.
 Fig.1 Workflow of spatial metabolomics. (Creative Biolabs)
Fig.1 Workflow of spatial metabolomics. (Creative Biolabs)
Published Data
| Paper Title | Spatially Resolved Multi-omics Highlights Cell-specific Metabolic Remodeling and Interactions in Gastric Cancer |
| Journal | Nature Communications |
| Published | 2023 |
| Abstract | Incorporating mass spectrometry imaging-based spatial metabolomics and lipidomics with microarray-based spatial transcriptomics, the study elucidates gastric cancer's intratumor metabolic heterogeneity and cellular metabolic interactions. By examining tumor-associated metabolic reprogramming at metabolic-transcriptional levels, marker metabolites, lipids, and genes are linked within metabolic pathways and co-localized amid diverse cancerous tissues. Spatial multi-omics integration cogently discerns cell types and distributions throughout the intricate tumor microenvironment, characterizing a pronounced immune cell-driven "tumor-normal interface" region where neoplastic cells engage neighboring tissues. This region exhibits distinct transcriptional patterns and substantial immunometabolic shifts. The innovative methodology for charting tissue molecular configurations yields a comprehensive perspective of intratumor disparities, revolutionizing comprehension of cancer metabolism on a systemic scale. |
| Result |
The study involved the examination of gastric cancer samples obtained from seven patients through a 10-micron frozen sectioning technique. Subsequent analyses employed advanced methodologies, including air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI) and matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) for spatial metabolomics (SM) and lipidomics (SL), respectively. Additionally, spatial transcriptomics (ST) utilizing 10x Genomics Visium at a resolution of 100 μm was performed on four samples. Distinct patterns of spatial expression disparities were observed in the clustering of metabolome and lipid profiles. A comprehensive unsupervised probabilistic latent semantic analysis (PLSA) segregated the variables into five fundamental components, reflecting the primary spatial characteristics of tissue metabolites and lipid species. Notably, the investigation of spatial transcriptomics identified ten discrete clusters within the gastric cancer tissue sections, each representing distinct spatial arrangements.
|
Features & Benefits
-
High-Resolution Metabolite Mapping:
Spatial metabolomics allows for high-resolution mapping of metabolites within tissue sections. This enables researchers to visualize the spatial distribution of metabolites and gain insights into metabolic processes in specific cellular contexts.
-
Integration with Multiomics:
Combining spatial metabolomics with other omics technologies, such as proteomics and transcriptomics, provides a comprehensive view of cellular function and interaction, enhancing the understanding of complex biological systems.
-
Artifact-Free Imaging:
Advanced technologies ensure virtually artifact-free imaging, providing accurate and reliable data for large-scale tissue analysis.
-
Enhanced Sensitivity and Specificity:
Innovations significantly improve the sensitivity and specificity of metabolite detection, allowing for the analysis of larger samples and more detailed metabolic profiling.
-
Hyperspectral Imaging:
The generation of hyperspectral imaging data enables the detection of biosynthesis and transport pathways of metabolites in tissues, which is key to understanding pathological mechanisms and developing targeted therapies.
Q&As
Q: How does Spatial Metabolomics benefit therapeutic research?
A: Spatial Metabolomics allows researchers to identify and map metabolic alterations in diseased tissues, such as cancer, providing insights into metabolic pathways and interactions within the tumor microenvironment. This can inform the development of targeted therapies by identifying metabolic vulnerabilities in cancer cells.
Q: What types of samples can be analyzed using Spatial Metabolomics?
A: Spatial Metabolomics can analyze a wide range of tissue types, including human tissues such as lung tumors and tonsil tissues. This flexibility makes it suitable for studying various diseases and conditions.
Q: Can Spatial Metabolomics be integrated with other omics techniques?
A: Yes, Spatial Metabolomics can be integrated with other omics techniques such as spatial transcriptomics and proteomics. This multi-omics approach provides a comprehensive view of the metabolic and molecular landscape of tissues, enhancing our understanding of cellular functions and interactions.
Q: How does Spatial Metabolomics contribute to drug development?
A: By providing detailed metabolic maps of tissues, Spatial Metabolomics helps identify biomarkers and therapeutic targets. Understanding the metabolic environment of diseased tissues can guide the development of drugs that target specific metabolic pathways, leading to more effective treatments.
Q: What makes Spatial Metabolomics unique compared to traditional metabolomics?
A: Unlike traditional metabolomics, which provides average metabolite concentrations across a sample, Spatial Metabolomics offers detailed maps of metabolite distribution, allowing for the study of metabolic heterogeneity and microenvironment-specific metabolic activities within tissues.
Resources
Reference
- Sun, Chenglong et al. "Spatially resolved multi-omics highlights cell-specific metabolic remodeling and interactions in gastric cancer." Nature communications vol. 14,1 2692. 10 May. 2023, doi:10.1038/s41467-023-38360-5. Distributed under Open Access license CC BY 4.0, without modification.
Search...


 Fig. 2 Spatially resolved multi-omics reveals intratumor heterogeneity of gastric cancer.1
Fig. 2 Spatially resolved multi-omics reveals intratumor heterogeneity of gastric cancer.1