Single Cell Epigenetic Service
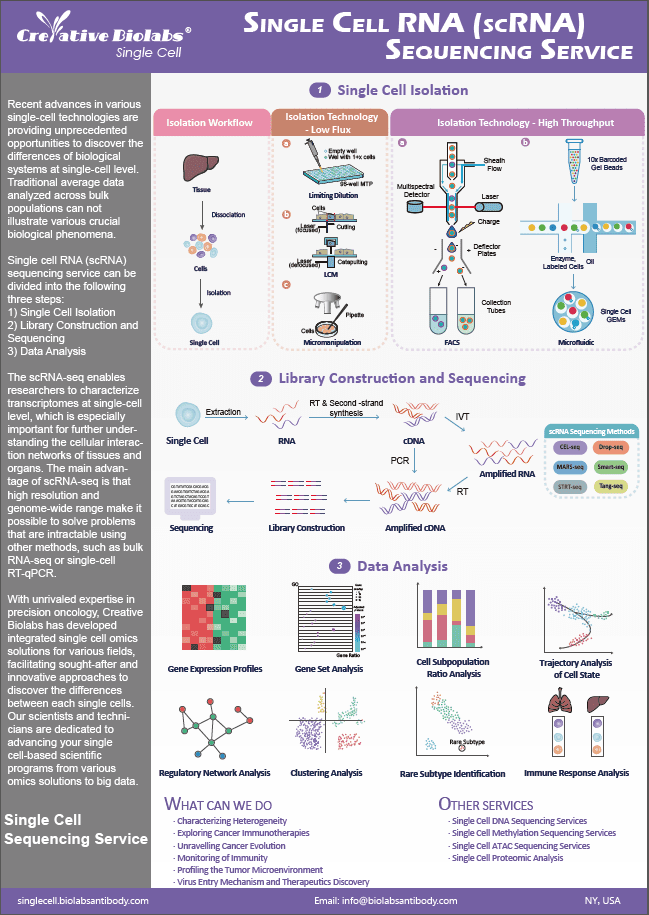
Our services
Creative Biolabs has validated progressive applications for single cell utility epigenetics. We allow for sample tracking during lab processing. Data QC, raw data, and interactive analysis reports are delivered securely.
Through covalent changes in histones and DNA, nucleosome position and density, substitution by histone variations, and epigenetic programming define cell phenotypes. While epigenetic changes are necessary for normal organism development, they have also emerged as hallmarks of cancer and other disorders in which specific cell types and individual cells play crucial roles. The finding of heterogeneity in cell populations at unprecedented resolution has been made possible by advances in single-cell genome transcriptome and DNA methylation research. It is possible to correlate genomic, transcriptomic, and epigenomic differences and dissect heterogeneity in complex tissues by combining multi-omic experiments inside the same cell.
 Fig.1 The principle of single cell epigenetic service.1.3
Fig.1 The principle of single cell epigenetic service.1.3
Single Cell Reduced Representation Bisulfite Sequencing
Creative Biolabs can assist you with every step of single-cell reduced representation bisulfite sequencing whether you wish to start with a small test project or require routine high-throughput services.
Learn MoreSingle Cell Whole Genome Bisulfite Sequencing
Creative Biolabs' single-cell whole-genome bisulfite sequencing service can examine single-cell DNA methylation to explain intercellular epigenetic diversity.
Learn MoreSingle Cell ChIP-Seq Service
Creative Biolabs offers single cell ChIP-seq for a better interpretation of the intra-tumor heterogeneity of chromatin states.
Learn MoreMethods of Single Cell Epigenetics Service
With methods for genome-wide mapping of DNA methylation, histone modifications, chromatin accessibility, and chromosome conformation, high-throughput sequencing has revolutionized the field of epigenetics. Initially, the input requirements for these methods necessitated samples containing hundreds of thousands or millions of cells; however, this has altered in recent years, with many epigenetic characteristics now being assayable at the single-cell level. Combined single-cell methods are also emerging that allow for the analysis of epigenetic–transcriptional correlations, allowing for more precise investigations into how epigenetic states are related to phenotypic.
 Fig.2 Epigenomics and the spectrum of single-cell sequencing technologies.2,3
Fig.2 Epigenomics and the spectrum of single-cell sequencing technologies.2,3
Applications of Single Cell Epigenomics
- To refine the understanding of epigenetic regulation
- To understand developmental processes and improve regenerative medicine
- To assess the complexity of cancer
Advantages of Our Service
- Assistance with experimental design and application selection based on analytical requirements, amount, and quality of input
- One-stop solutions including extraction, sample QC, library preparation, sequencing, and bioinformatics analysis
- Advanced analysis offering includes custom visuals and publication-ready figures for improved data interpretation
- Comparative analysis with other epigenomic data such as ChIP seq, ATAC seq, FAIRE seq, and RNA seq data
Results Display
Here are some results displayed that refer to the drawings of single cell epigenomics articles
- t-SNE analysis of single-cell gene expression and DNA methylation data.
- sc-GEM analysis of single-cell gene expression and DNA methylation. The top color bar indicates the genotype of each cell. Hierarchical clustering of single cells according to gene expression profiles and DNA methylation profiles yielded two clusters.
- Combined expression (snDrop-seq) data showing distinct cell-type and subtype clustering visualized by t-distributed stochastic neighbor embedding.
- A heat map of selected genes. Columns represent data sets ordered by the pseudotime trajectory.
Creative Biolabs is a dependable service provider to offer single cell epigenetic services for our global customers. Please contact us for more information about our single cell epigenetic services.
Q&As
Q: What are the main applications of Single Cell Epigenetic Service?
A: The main applications include understanding epigenetic regulation, studying developmental processes, improving regenerative medicine, and assessing the complexity of cancer. This service helps in uncovering cell heterogeneity and identifying biomarkers and therapeutic targets.
Q: What techniques are used in Single Cell Epigenetic Service?
A: Techniques include single-cell whole-genome bisulfite sequencing (scWGBS), single-cell reduced representation bisulfite sequencing (scRRBS), single-cell chromatin immunoprecipitation sequencing (scChIP-seq), and single-cell ATAC-seq for chromatin accessibility.
Q: How does single-cell ChIP-seq contribute to epigenetic research?
A: Single-cell ChIP-seq (scChIP-seq) maps histone modifications and transcription factor binding sites at a single-cell level. This technique is essential for studying chromatin states and their role in gene regulation and disease mechanisms.
Q: What are the advantages of using Single Cell Epigenetic Service?
A: Advantages include high-resolution epigenetic profiling, the ability to analyze rare and heterogeneous cell populations, comprehensive data analysis, and customized bioinformatics support. These benefits facilitate precise investigations into gene regulation and disease.
Q: How can Single Cell Epigenetic Service aid in cancer research?
A: This service helps in understanding cancer cell heterogeneity, identifying epigenetic changes associated with metastasis and treatment resistance, and discovering new biomarkers and drug targets. It provides insights into cancer cell plasticity and evolution.
Resources
References
- Tang, Xiaoning et al. "The single-cell sequencing: new developments and medical applications." Cell & bioscience vol. 9 53. 26 Jun. 2019, doi:10.1186/s13578-019-0314-y.
- Clark, Stephen J et al. "Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity." Genome biology vol. 17 72. 18 Apr. 2016, doi:10.1186/s13059-016-0944-x.
- Distributed under Open Access license CC BY 4.0, without modification.
Related Sections
Search...