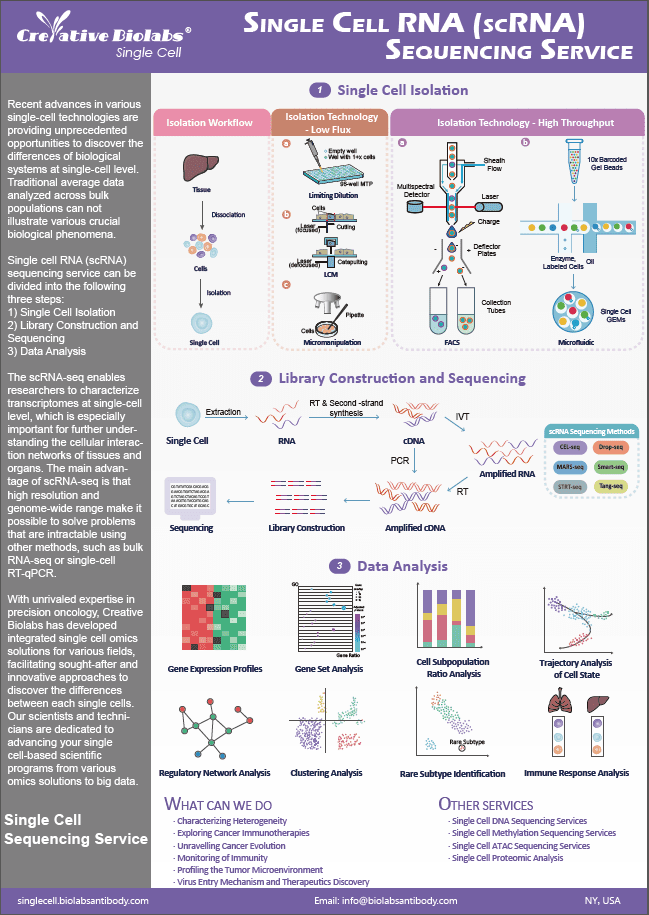
Single Cell TCR Profiling Service
Creative Biolabs provides a comprehensive range of customized, high-quality services in single cell TCR profiling service to support scientific research in immunology and related biomedical industry worldwide, with proven experience and expertise in exploring immune receptor mapping and discovering new biomarkers.
TCR
The peculiarity of T cell is their ability to recognize an infinite range of self and foreign antigens. This ability is achieved during thymic development through a complex molecular mechanism based on somatic recombination that leads to the expression of a very heterogeneous population of surface antigen receptors, the T Cell Receptors (TCRs).
TCR is composed of two different protein chains. The vast majority of T cells express TCRs composed of α (alpha) and β (beta) chains while a small subset expresses TCR composed of γ (gamma) and δ (delta) chains. The genes encoding alpha (TCRA) and beta (TCRB) chains are composed of multiple noncontiguous gene segments which include variable (V), diversity (D), and joining (J) segments for TCRB gene and variable (V) and joining (J) for TCRA gene. The enormous diversity of T cell repertoires is generated by random combinations of germ line gene segments (combinatorial diversity) and by random addition or deletion at the junction site of the segments that have been joined (junctional diversity). (Schatz and Ji, 2011)
 Fig.1 Somatic V(D)J arrangement in the alpha and beta chains. (A) Genomic organization and somatic recombination of TCRB and TCRA loci. (B) Productive arrangements of beta and alpha transcripts. (C) Organization of TCR.1, 3
Fig.1 Somatic V(D)J arrangement in the alpha and beta chains. (A) Genomic organization and somatic recombination of TCRB and TCRA loci. (B) Productive arrangements of beta and alpha transcripts. (C) Organization of TCR.1, 3
Single Cell TCR Profiling
Since the launch of 10x genomics platform in 2015, single cell technology can provide sequence information on paired alpha and beta chains of individual cells. Single T cells are encapsulated into droplets, where they undergo OE-PCR to enable linking of TCRα and TCRβ transcripts. Fused TCRαβ transcripts are then pooled, amplified and sequenced.
Our Single Cell TCR Profiling Service
We offer scientific and meticulous design for sample preparation, library construction, sequencing and data analysis to ensure high-quality research results. For more information about immune repertoire sequencing, please inquire our single cell immune profiling service.
 Fig.2 Our Single Cell TCR Profiling Service. (Creative Biolabs)
Fig.2 Our Single Cell TCR Profiling Service. (Creative Biolabs)
Features & Benefits
Creative Biolabs is committed to efficiency, confidentiality, and rapid, accurate service with proven experience and expertise.
 Fig.3 Our features and benefits. (Creative Biolabs)
Fig.3 Our features and benefits. (Creative Biolabs)
Sample Requirements
- Sample type: fresh human or mouse T cell suspension, blood B cell suspension
- Quality requirements: Cell viability is greater than 90%, concentration is 500-2000 cell/μL
- Cell size: 5 um ≤ X ≤ 30um
Before delivering cells, please contact us to discuss the sample preparation.
Results Display
Here are some results displayed that refers to the drawings of single cell T cell receptor sequencing profiling articles.
- VDJ gene display map for pairing immune cell receptor
- UMAP plot
- Customized data analysis
 Fig.4 UMAP visualization of T cells.2, 3
Fig.4 UMAP visualization of T cells.2, 3
 Fig.5 The proportion of unique and non-unique TCRs expression in interested T cells.2, 3
Fig.5 The proportion of unique and non-unique TCRs expression in interested T cells.2, 3
 Fig.6 Applications of single cell TCR profiling. (Creative Biolabs)
Fig.6 Applications of single cell TCR profiling. (Creative Biolabs)
Data analysis can be customized based on the needs of customers.
Applications
Single cell technology can provide sequence information on paired alpha and beta chains of individual cells. A further level of complexity derives by the efforts to associate TCR repertoire and gene expression profile from the same cells. This kind of analysis provides an unbiased classification of a population of interest and the association of the transcriptional landscape of each cell with its TCR. This approach promises to open new avenues to describe the specific clonality of immune cell subsets even of the poorly characterized phenotypic subtypes and gives the opportunity to monitor the effect of transcriptional dynamics in strict association to clonality, which can help researchers explore T cell development and differentiation and have a wide range of applications in autoimmunity and cancer immunotherapy.
 Fig.7 Applications of single cell TCR profiling. (Creative Biolabs)
Fig.7 Applications of single cell TCR profiling. (Creative Biolabs)
Please contact us for your tailored solution.
Q&As
Q: How does the Single Cell TCR Profiling Service work?
A: The service involves isolating single T cells, extracting RNA, and sequencing the TCR alpha and beta chains using high-throughput sequencing technologies. This process generates detailed TCR profiles for each cell, enabling the study of T cell repertoire diversity and dynamics.
Q: How does Single Cell TCR Profiling benefit cancer research?
A: By profiling TCRs at the single-cell level, researchers can identify tumor-infiltrating lymphocytes and understand their role in tumor immunity. This information helps in developing targeted immunotherapies and predicting patient responses to treatment.
Q: How does Single Cell TCR Profiling aid in autoimmune disease research?
A: This service helps in identifying autoreactive T cells and understanding their role in disease progression. Profiling the TCR repertoire in autoimmune diseases can reveal mechanisms of autoimmunity and guide the development of targeted treatments.
Q: How does Single Cell TCR Profiling contribute to vaccine development?
A: By analyzing the TCR repertoire in response to vaccination, researchers can identify T cell clones that are activated by the vaccine. This information helps in understanding vaccine efficacy and guiding the design of more effective vaccines that elicit strong and durable immune responses.
Q: How does the platform handle low-abundance TCR clones?
A: The platform employs sensitive sequencing techniques and robust data analysis pipelines to detect low-abundance TCR clones. This capability is essential for capturing the full diversity of the T cell repertoire, including rare clones that may be critical for immune responses.
Resources
References
- De Simone, Marco et al. "Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges." Frontiers in immunology vol. 9 1638. 18 Jul. 2018, doi:10.3389/fimmu.2018.01638.
- Yang, Hu-Qin et al. "Single-Cell TCR Sequencing Reveals the Dynamics of T Cell Repertoire Profiling During Pneumocystis Infection." Frontiers in microbiology vol. 12 637500. 20 Apr. 2021, doi:10.3389/fmicb.2021.637500.
- Distributed under Open Access license CC BY 4.0, without modification.
Search...