Protein Microarray Service
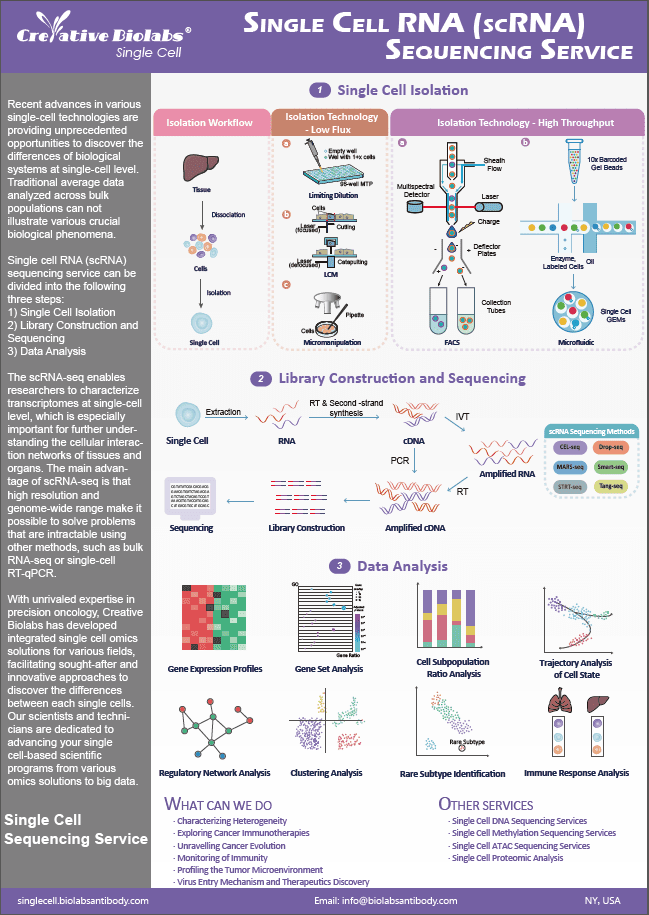
Protein arrays technology was developed in the early 2000s to do multiplexed protein measurements to explore protein function, protein-protein interactions, and modeling networks and pathways with the same throughput as DNA arrays.
Protein Microarray
Protein microarray, also known as protein chips, is a recently developed proteomic tool for characterizing the interactions, functions, and activities of proteins in a high-throughput manner. These tests can be scaled down to fit on chips designed for running on protein microarrays. The ability of protein microarrays to examine interactions between the nucleus and proteins, as well as interactions between proteins themselves, as well as interactions between ligands and receptors, drug targets, and protein substrates on a large scale, is one reason for their growing popularity. Briefly, protein microarrays are classified as target microarrays, and in situ expressed arrays.
- Target Microarray
The most frequent type of target microarray is an antibody microarray. Target microarrays are made up of various proteins printed onto a planar-solid surface. Since they allow the detection of specific protein profiles, these arrays have been found in various applications in clinical studies.
- In situ Expressed Array
In situ expressed protein arrays are characterized for in vivo transcription-translation protein expression at the moment of the assay by using cell-free expression systems such as Escherichia coli, HeLa cell lysates, or rabbit reticulocyte lysates. This array requires a complementary DNA clone collection, which codifies proteins of interest and tags in the carboxy or amino terminus.
Creative Biolabs offers different panels of antibody combinations for the specific analysis of different biological processes, including cardiovascular disease panel, oncology panel, inflammation panel, nervous system panel, immuno-oncology transition panel, cell cycle regulation panel, cardiac metabolism panel, development biology panel, immune response panel, metabolism panel, neural exploration panel, and organ damage panel. For more information, please contact us.
Features & Benefits
-
High-throughput Analysis
Protein microarrays allow simultaneous analysis of thousands of proteins. This high-throughput capability accelerates the identification of potential drug targets and biomarkers, crucial for advancing therapeutic research and development. -
Rapid Results
Protein microarrays provide results faster than traditional methods, facilitating quicker decision-making in the drug development process. This speed is vital for meeting critical project timelines in fast-paced research environments. -
Low Sample Volume Requirement
These arrays require only small amounts of sample material, preserving precious research samples and allowing for the study of limited or rare specimens, which is particularly beneficial in clinical research settings. -
Compatibility with Diverse Sample Types
Protein microarrays can analyze a variety of sample types including blood, serum, plasma, and tissue extracts, offering flexibility in experimental design and the ability to study complex biological matrices. -
Cost-Effective
Compared to other high-throughput technologies like mass spectrometry, protein microarrays are more cost-effective, allowing for broader experimental designs within budget constraints.
Q&As
Q: What types of protein microarrays are available?
A: There are several types of protein microarrays including analytical, functional, and reverse-phase protein arrays. Each type serves different purposes: analytical arrays for detecting protein presence, functional arrays for studying protein interactions, and reverse-phase arrays for observing protein abundance from complex samples.
Q: What sample types can be used with protein microarrays?
A: Protein microarrays can analyze a variety of sample types including serum, plasma, tissue extracts, and cell culture supernatants. This versatility allows researchers to conduct studies across different biological matrices, enhancing the applicability of the microarrays in clinical and research settings.
Q: How sensitive are protein microarrays?
A: Protein microarrays offer high sensitivity, comparable to or better than traditional ELISA tests. This high sensitivity makes them suitable for detecting low-abundance proteins and subtle changes in protein activity, crucial for understanding disease mechanisms or evaluating drug effects.
Q: Can protein microarrays be used for quantifying protein modifications?
A: Yes, protein microarrays are particularly useful for profiling post-translational modifications (PTMs) such as phosphorylation, glycosylation, and ubiquitination. This capability is crucial for understanding protein function in different cellular contexts and disease states, offering insights into cellular signaling pathways and potential therapeutic targets.
Q: What support and resources are available for protein microarray users?
A: Many providers of protein microarray services offer extensive support and resources, including detailed protocols, troubleshooting guides, and technical assistance. These resources are invaluable for users new to the technology or those encountering specific issues with their experiments.
Resources
Search...