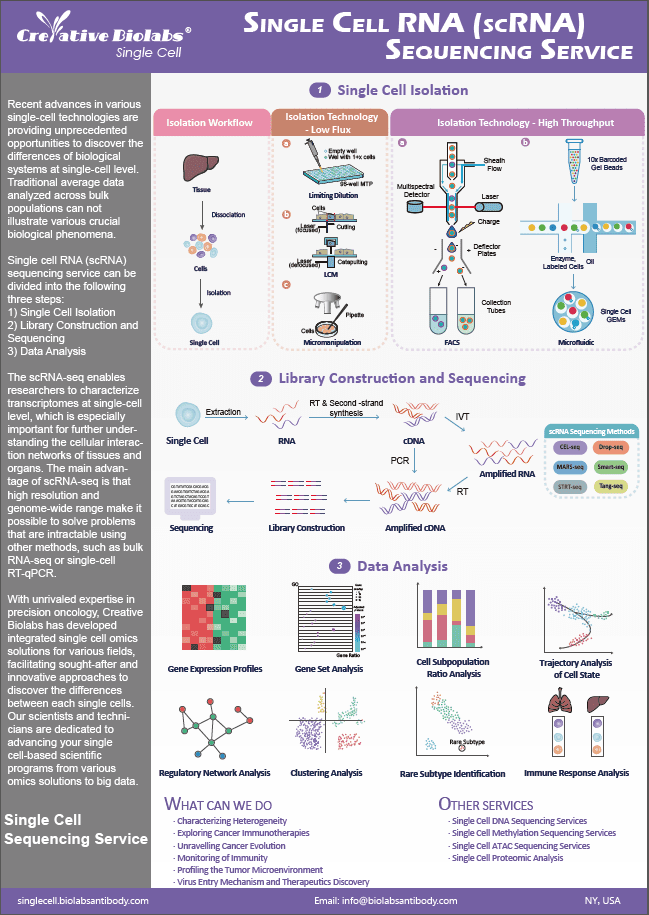
Single Cell Target DNA Sequencing Service
Conventional bulk sequencing techniques are inadequate for detecting the genetic diversity that exists across cell populations. The single cell target DNA sequencing services we provide offer an accessible and rapid means of detecting genomic variability within and across cell populations. Our technology is well-suited to high-impact application areas such as blood cancers, solid tumors, and genome editing validation. Our innovative approach, which utilizes targeted gene panels for single-cell DNA analysis, provides a powerful means of detecting rare subclones, resolving mutational co-occurrence patterns, determining zygosity, and reconstructing phylogenetic lineages. For single cell genomics to have a real impact on personalized medicine and inform clinical decision-making, it is imperative that it be sensitive, accurate, and capable of detecting low-abundance clones.
Applications of Microdroplet-Based Single Cell Target DNA Sequencing
- Identify rare cell populations.
- Co-occurring mutations to identify.
- Count the zygosity.
- Clarify the clonal heterogeneity.
Benefits of Microdroplet-Based Single Cell Target DNA Sequencing
- From thousands of single cells, identify SNVs, indels, CNVs, LOHs, and translocations.
- Combine with protein panels to simultaneously learn about single-cell phenotypes and genotypes.
- Use targeted panels that are focused on your genes or regions of interest to take advantage of the flexibility in experimental design and budget.
- Create your own DNA and protein panels for hematology or solid tumors or select from pre-designed panels.
 Fig.1 Microdroplet-based single cell target DNA sequencing workflow. 1
Fig.1 Microdroplet-based single cell target DNA sequencing workflow. 1
Sample Requirements
- Frozen single cell suspensions: ≥ 5x106 cells per sample, viability ≥ 80%.
- Fresh PBMC samples: use special PBMC separation tubes, or ordinary EDTA anticoagulation tubes to collect blood (8-10 ml), centrifuge, and separate PBMC within 2 hours after blood collection. Separated PBMC is added to the tissue preservation solution and transported in ice bags.
- Fresh peripheral blood samples: Blood collection in normal EDTA anticoagulation tubes (8-10 ml), transported in ice bags.
- Fresh bone marrow samples: added to tissue preservation solution after sampling, and transported in ice packs.
- Cell size: < 40 μm in diameter.
Workflow of Single Cell Target DNA Sequencing Service
 Fig.2 Basic workflow of single cell DNA sequencing. (Creative Biolabs)
Fig.2 Basic workflow of single cell DNA sequencing. (Creative Biolabs)
Bioinformatics Analysis
- Remove joint contamination sequence and low-quality data
- Matching, output data statistics
- SNP detection, annotation, and statistics
- InDel detection, annotation, and statistics
- CNV detection, annotation, and statistics
- SV detection, annotation, and statistics
- Customized information analysis: Customers can negotiate the content of customized information analysis, such as group structure analysis, principal component analysis, and phylogenetic tree construction.
Published Data
| Paper Title | Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing |
| Journal | Cell |
| Published | 2018 |
| Abstract | In this study, they profiled longitudinal samples from 20 Triple-Negative Breast Cancer (TNBC) patients during neoadjuvant chemotherapy (NAC) using single-cell DNA and RNA sequencing, as well as bulk exome sequencing. Deep exome sequencing revealed ten patients in whom NAC resulted in clonal extinction and ten patients in whose clones survived therapy. They did a more extensive investigation on 8 patients, analyzing 900 cells with single-cell DNA sequencing and 6,862 cells with single-cell RNA sequencing. |
| Result | They looked at the genomic and phenotypic evolution of tumor cells in TNBC patients as a result of NAC treatment and discovered two types of clonal dynamics: extinction and persistence. After treatment, NAC eradicated the tumor cells, leaving only normal diploid cell types, including numerous fibroblasts and immunological cells, in the clonal extinction patients. The clonal persistence patients, on the other hand, had a substantial number of remaining tumor cells with altered genotypes and phenotypes in response to NAC. |
Creative Biolabs' Service
Creative Biolabs provides microdroplet-based single cell target DNA sequencing services, which enable high-throughput analysis of specific gene regions of interest. This approach is ideal for exploring cell populations and uncovering genetic variation at the single cell level. It is important to note that this service must specify a detection panel.
Please do not hesitate to contact us for more details.
Q&As
Q: What are the primary applications of this service?
A: This service is primarily used in cancer research to identify rare subclones, assess mutational co-occurrence, determine zygosity, and reconstruct phylogenetic lineages. It is also valuable in genome editing validation, studying circulating tumor cells, and understanding tumor microenvironments.
Q: What sequencing technologies are used?
A: This service typically uses advanced sequencing platforms such as Illumina NovaSeq and Mission Bio Tapestri system, which offer high throughput and accuract for single-cell sequencing.
Q: How does this service support cancer research?
A: It helps in identifying genetic mutations associated with cancer, analyzing tumor heterogeneity, understanding the tumor microenvironment, and discovering new therapeutic targets. It is particularly useful in researching hematological tumors, breast cancer, brain tumors, and head and neck squamous cell carcinoma.
Q: Can this service be used to study the immune response in diseases?
A: Yes, single-cell sequencing can map immune cell populations and their gene expression profiles in various diseases, including cancer and infectious diseases like COVID-19. This helps in understanding immune cell heterogeneity and discovering immune-related biomarkers
Q: What are the bioinformatics capabilities of this service?
A: The bioinformatics pipeline includes quality control, sequence alignment, mutation detection, and comprehensive statistical analysis. Customizable analysis options allow researchers to tailor data outputs to specific research questions, enhancing the depth of genomic insights.
Resources
Reference
- Sun, Weiwei et al. "Phenotypic signatures of immune selection in HIV-1 reservoir cells." Nature vol. 614,7947 (2023): 309-317. doi:10.1038/s41586-022-05538-8. Distributed under Open Access license CC BY 4.0, without modification.
Search...