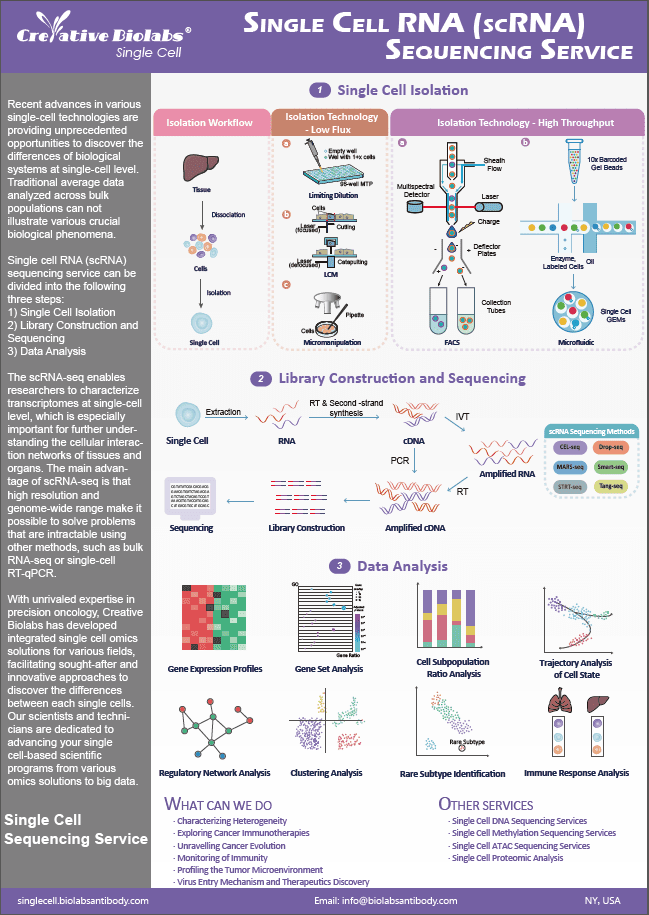
Exploring Cancer Immunotherapies
The development of cancer immunotherapies relies on an accurate understanding of the cellular composition and complex interactions between cells in the tumor microenvironment. To discover the secrets inside individual cells, Creative Biolabs developed a comprehensive suite of single cell omics solutions for your immuno oncology programs. We are dedicated to assisting you with exploring insights for novel cancer immunotherapies.
Cancer Immunotherapy
Traditional cancer treatments include surgery, radiation, and chemotherapy. In recent years, cancer immunotherapy has achieved major success in the fight against cancer, through deliberate and specific enhancement of the natural immune system. Both active and passive immunotherapies have proven efficacious against various cancer types by increasing the anti-cancer immune capabilities or limiting the tumor evasion. Chimeric antigen receptor (CAR) therapy is active immunotherapy aimed at educating the patients' immune system to recognize and fight cancer. Checkpoint inhibitors are passive immunotherapy aimed at attenuation of inhibitory T cells, leading to the active status of effector cancer-fighting T cells. However, tumor research is beset by cellular heterogeneity. Nowadays, single-cell techniques have emerged as an advanced solution for analyzing tumor heterogeneity. Next-generation sequencing systems make it possible to sequence DNA or RNA at single-cell level. With extensive experience in single cell omics, we can accelerate your cancer immunotherapy program and maximize your success with single-cell data-driven precision research.
Identifying Novel Targets for Immunotherapies
In a recent study, the researchers analyzed a large number of scRNA-seq data to identify intratumoral heterogeneity of the immune response gene. Studies have shown that the expression of the IFN-γ signaling pathway gene is heterogeneous in cancer cells and is regulated by certain genes, including MHC class II genes. In addition, down-regulation of genes in the IFN-γ signaling pathway corresponds to acquired resistance. They also discovered the heterogeneity of expression of new antigens and cancer/testis antigen (CTA). The heterogeneity of IFN-γ expression may be one of the reasons for poor treatment.
Identifying Novel Prognostic Indicators
Single cell genomics can be used to identify new prognostic indicators. The researchers used melanoma patients treated with checkpoint inhibitors to analyze the transcriptome of 16,291 immune cells from 48 tumor samples. They point out that although about 40% of melanoma patients respond to PD-1 immunological checkpoint therapy, most patients either do not respond or relapse after treatment. They used scRNA-seq to analyze immune cells and correlated the cell transcriptome with patient history and clinical status. The study identified the status of several CD8+ T cells associated with melanoma lesions. They found that the presence of TCF7 protein in CD8+ cells predicts whether patients respond to checkpoint inhibitor therapy and is associated with a better prognosis. This information helps predict which patients may benefit from checkpoint blocking therapy.
Insights for Combinatorial Immunotherapies
Single-cell genomics, publicly accessible databases, and global scientific collaborations are driving research for cancer immunotherapy. As the proportion of single-cell data continues to increase, these resources will become very valuable. With rich cooperation resources, feasible multi-omics methods and practical single-cell analysis programs, the problem of tumor heterogeneity will be solved, and the research on cancer immunotherapy is expected to achieve another milestone. The single-cell data can drive the development of combinatorial immunotherapies in a more precise way.
 Fig.1 Process for single cell RNA-seq and cancer cell identification in low grade brain tumors.1
Fig.1 Process for single cell RNA-seq and cancer cell identification in low grade brain tumors.1
Identifying Co-occurring Mutations
Tumor tissue blocks can show the genetics of the tumor, but important differences between tumor cells are masked. According to histopathological and molecular genetic studies, IDH-mutated astrocytoma (IDH-A) and oligodendroglioma (IDH-O) are completely different in genetics and histopathology. However, the markers GFAP and OLIG2 of astrocytes and oligodendrocytes can be found in both tumor cells, suggesting that these two tumors are not completely different in the cell ancestry. By single-cell sequencing of astrocytoma and oligodendroglioma, and comparing with cancer genome mapping (TCGA) data, the scientists found that both tumors share a common glioma progenitor cell. IDH-A and IDH-O have common undifferentiated primitive cells or neural precursor cells, the main difference between which is caused by genetic variation and tumor microenvironment. Induction of precursor cell differentiation may hinder the growth of tumor cells.
With unrivaled expertise and experiences in end-to-end single cell omics solutions, Creative Biolabs offers integrated solutions for the entire program from sample preparation, library construction, sequencing, to data analysis. You can partner with us for the entire program or just one module. We offer you complete flexibility. To explore more details, please contact us for assistance.
Features & Benefits
-
Comprehensive Immunoprofiling
Provides detailed immunoprofiling of the tumor microenvironment to identify key players in the immune response against cancer.
-
Identification of Neoantigens
Identifies tumor-specific neoantigens that can be targeted by personalized cancer vaccines or T-cell therapies, enhancing the specificity and efficacy of immunotherapies.
-
Checkpoint Inhibitor Analysis
Analyzes the expression of immune checkpoint molecules to predict patient response to checkpoint inhibitor therapies and guide treatment decisions.
-
Tumor-Immune Interactions
Investigates interactions between tumor cells and immune cells, revealing mechanisms of immune evasion and potential therapeutic targets to overcome resistance.
-
Single-Cell Resolution
Utilizes single-cell technologies to examine the heterogeneity of immune cell populations within tumors, providing insights into the diverse immune landscape and its impact on therapy.
Q&As
Q: How does this service help identify potential therapeutic targets?
A: By profiling immune cell populations and their interactions with tumor cells, the service identifies key molecules and pathways that can be targeted to enhance anti-tumor immune responses.
Q: What types of immunotherapies can be developed using insights from this service?
A: Insights from this service can be used to develop a range of immunotherapies, including checkpoint inhibitors, CAR-T cell therapies, cancer vaccines, and combination therapies.
Q: Can this service predict patient response to immunotherapy?
A: Yes, by analyzing the expression of immune checkpoint molecules and other biomarkers, the service helps predict which patients are likely to respond to specific immunotherapies.
Q: What role does cytokine profiling play in this service?
A: Cytokine profiling helps understand the inflammatory environment within tumors, revealing how cytokines influence immune cell function and the overall immune response to cancer.
Q: How does the service handle the analysis of rare immune cell populations?
A: Advanced single-cell and high-throughput technologies are used to detect and analyze rare immune cell populations, providing detailed insights into their roles and contributions to the tumor immune microenvironment.
Resources
Reference
- Reitman, Zachary J et al. "Mitogenic and progenitor gene programmes in single pilocytic astrocytoma cells." Nature communications vol. 10,1 3731. 19 Aug. 2019, doi:10.1038/s41467-019-11493-2. Distributed under Open Access license CC BY 4.0, without modification.
Search...