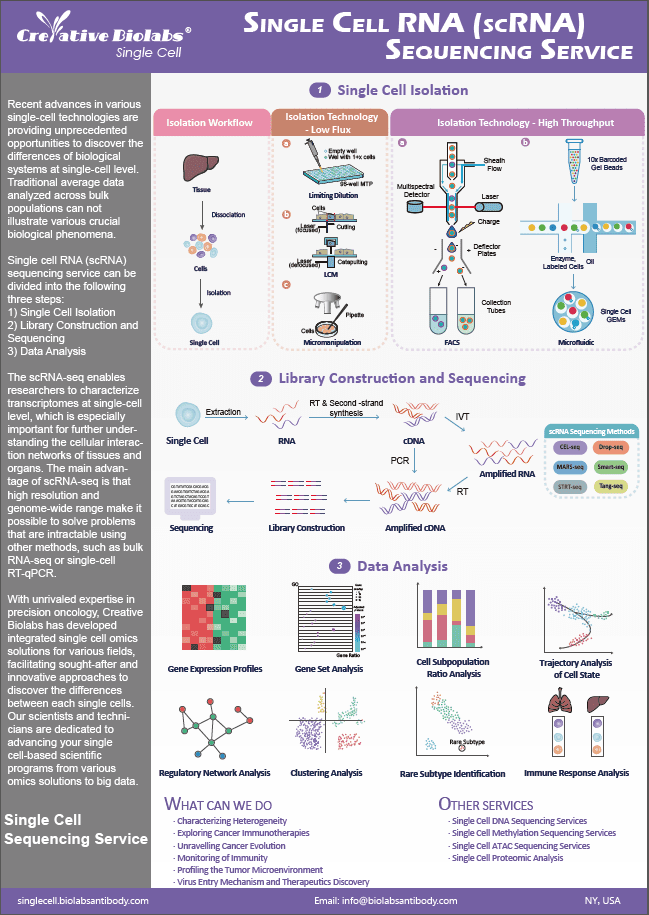
- Home
- Single Cell Research Area
- Single Cell Omics Solutions for Immunity and Microbiome
- Single Cell Omics Solutions for Immune Cells
- Single Cell Omics Solutions for Natural Killer T (NKT) Cells
Single Cell Omics Solutions for Natural Killer T (NKT) Cells
Natural Killer T (NKT) cells from different lymphocyte lineages at the interface between innate and adaptive immunity, but their role in immune response remains elusive. Therefore, it is very important to make a comprehensive analysis of NKT cells. Whether you want to dissect differences in cell types, study adaptive immune systems, or discover copy number variations and genomic heterogeneity, Creative Biolabs's various analytical platforms based on single-cell omics are able to answer your scientific questions.
Single-Cell Genome Analysis for NKT Cells
The analysis of gene expression patterns and genome analysis has become a useful tool for studying the biological functions of different cell types and characterizing functional distributions that distinguish between distinct phenotypes or stages of differentiation or activation. We analyzed the gene expression profiles of NKT cells in order to obtain basic information about the function of NKT cells in congenital and adaptive immune responses. In addition, resting and activated NKT cells were analyzed to reveal their functional plasticity in immune response. Our findings reveal unique gene expression profiles and multiple effector functions of resting NKT cells compared with Natural Killerer (NK) cells, naïve conventional CD4+ T helper (Th) and regulatory T cells (Treg), which not only activate known Th1 and Th2 phenotypes but also connect to Th17 phenotypes.
Single-Cell Transcriptomic Analysis for NKT Cells
Publicly available evidence on single-cell transcriptomes suggests that there may be substantial differences between individual cells in the "same" population, which may have important implications for understanding the origin and function of the population. single-cell RNA sequencing (RNA-Seq) analysis of NKT cells showed that NKT cells could serve as a bridge between innate and adaptive immunity mediating immune response. Unlike NK cells or T cells, NKT cells exhibit unique tissue specificity under steady-state conditions, suggesting that the function of strip segmentation is more meaningful for further study of diseases.
Single-Cell Epigenetic Analysis for NKT Cells
Epigenetic modification has been shown to be the key to control normal hematopoiesis, such as the consequences of changes in DNA methylation in primitive hematopoietic cells that are destroyed from the beginning. Epigenetics analysis was performed by microarray analysis of chromatin to understand the mechanism of gene regulation of NKT cell function. Our findings provide new insights into the NKT cell family, which may guide further research to better manipulate NKT cells for therapeutic applications.
Highlights
- Detection ability of large-scale samples
- High sensitivity, no preference
- Accurate analysis process
Creative Biolabs is dedicated to enhancing your biological discoveries at single-cell level. We are confident in unraveling secrets in individual cells. If you want to know more about our single cell omics solutions, please feel free to contact us for more details.
Q&As
Q: Can single-cell omics be used to study NKT cell activation?
A: Yes, single-cell omics technologies like single-cell proteomics and methylation sequencing can be used to study the activation states of NKT cells. These methods help dissect the molecular mechanisms behind NKT cell activation and their subsequent effects on other immune cells.
Q: What types of omics data can be generated from Single Cell Omics for NKT cells?
A: This service can generate various types of omics data, including single-cell RNA sequencing (scRNA-seq) for gene expression, single-cell ATAC sequencing (scATAC-seq) for chromatin accessibility, and single-cell proteomics for protein expression profiles. These data types provide a multidimensional view of NKT cell biology.
Q: How can Single Cell Omics Solutions advance NKT cell research?
A: This service advances NKT cell research by providing high-resolution data on individual cell states and interactions. It helps researchers identify rare NKT cell subpopulations, understand cell-specific responses to stimuli, and uncover regulatory mechanisms driving NKT cell functions.
Q: What expertise is required to perform Single Cell Omics on NKT cells?
A: Performing Single Cell Omics on NKT cells requires expertise in immunology, single-cell technologies, and bioinformatics. Researchers need to be skilled in cell isolation, library preparation, sequencing, and data analysis to ensure high-quality, interpretable results.
Q: How can single-cell omics data be integrated with other types of biological data to enhance NKT cell research?
A: Integrating single-cell omics data with other biological data, such as bulk sequencing data, clinical data, and imaging data, can provide a more comprehensive understanding of NKT cell function. This multi-dimensional approach allows researchers to correlate molecular profiles with phenotypic characteristics and clinical outcomes, leading to more robust conclusions and potential therapeutic applications.
Resources
Search...